More than a year after the World Health Organization (WHO) declared the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) outbreak a Public Health Emergency of International Concern, coronavirus disease 2019 (COVID-19) has caused more than 3.7 million deaths.
(1) Globally, public health systems are severely impacted and are further challenged by the emergence of SARS-CoV-2 variants carrying mutations that are of concern (VOC).
(2) Molecular diagnostic tools, particularly reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for viral RNA detection and next-generation sequencing (NGS) for molecular monitoring SARS-CoV-2 genetic diversity at the whole genome level, have proven critical for public health decision-making.
(3) Investigating SARS-CoV-2 genomes by NGS to track transmission chains, understand transmission dynamics, and rapidly identify mutations that potentially have an impact on transmissibility, morbidity, and mortality, as well as potential escape of diagnostic tools or vaccine-induced immunity have become an integral part of public health measures during this pandemic.
(4)Since the first whole genome sequence (WGS) analysis of SARS-CoV-2 has been published in January 2020,
(5) the virus has been continuously sequenced, characterized, and data made publicly available through global initiatives such as Global Initiative on Sharing All Influenza Data (GISAID). More than 1.8 million SARS-CoV-2 sequences have been publicly shared via GISAID, and numerous mutations in the gene encoding the spike protein have been identified.
(6) For example, the D614G variant has been shown to increase the viral load of infected patients and has replaced the original variant since June 2020 around the globe.
(7) More recently, SARS-CoV-2 lineages characterized by a combination of multiple mutations in the spike gene have emerged independently in different regions of the world. The SARS-CoV-2 lineages B.1.1.7 (also known as the Alpha variant, VOC 202012/01 or 501Y.V1), B.1.351 (also known as the Beta variant or 501Y.V2), and P.1 (also known as the Gamma variant, B.1.1.28.1 or 501Y.V3) were the first VOCs identified.
(8) The Alpha variant (lineage B.1.1.7) was first described in mid-December 2020 in the United Kingdom, and the mutation appears to have substantially increased transmissibility and has quickly developed into the dominant variant circulating in the UK and beyond.
(9) The Beta variant (lineage B.1.351) was identified in December 2020, which emerged most likely in South Africa and is also associated with higher transmissibility.
(10) The Gamma variant (lineage P.1) was identified in January 2021 in Manaus, the largest city in the Amazon region of Brazil.
(11) In January 2021, this region experienced a resurgence of COVID-19 despite the reported high seroprevalence of antibodies against SARS-CoV-2 in this population.
(12,13) During the preparation and revision of this manuscript, the WHO had designated the emerging lineage B.1.617.2 as the Delta VOC.
To rapidly detect and continuously monitor the appearance, introduction, and spread of (novel) VOCs, the level of molecular surveillance needs to be increased globally. The gold standard of genomic surveillance, NGS, allows for unbiased identification of mutations, but is limited by its relatively slow sample-to-result turnaround time and level of laboratory infrastructure and scientific expertise required. Furthermore, the relatively high costs of NGS increase the financial burden on establishing a widespread VOC-tracking strategy, a limiting factor particularly for resource-limited settings. Therefore, mutation-specific PCR-based approaches that are more cost-efficient and allow for high-throughput screening of a significant proportion of SARS-CoV-2-positive individuals were developed.
(14,15) To identify the transmission dynamics of VOCs in settings with limited sequencing capabilities, we have designed rapid and cost-efficient RT-qPCR assays detecting relevant mutations in the spike protein of SARS-CoV-2. The N501Y mutation is found in the Alpha, Beta, and Gamma VOCs, while the E484K mutation is restricted to Beta and Gamma variants. The Alpha variant is characterized by an additional spike gene deletion (ΔHV69/70). To further simplify, decentralize, and speed up the process of VOC identification, we transformed our assay to a portable and inexpensive qPCR device, named
peakPCR.
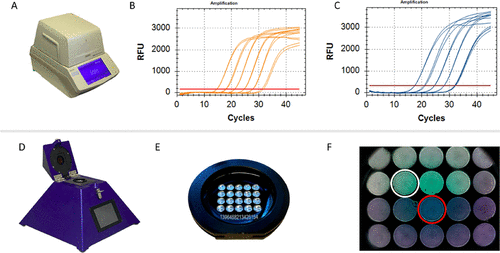
(16) The device can complete up to 20 RT-qPCR reactions in less than 40 min. Important characteristics of
peakPCR are the relatively low cost of production and the simplicity of usage. By using cartridges that are preloaded with lyophilized RT-qPCR reagents, the user interaction is reduced to loading the sample onto the cartridge. Furthermore, the preloaded
peakPCR cartridges can easily be shipped and stored at room temperature, making cold chains superfluous. Here, we report the development of a new approach for rapid, robust, and decentralized identification of SARS-CoV-2 VOCs that can be both run on standard laboratory RT-qPCR equipment and the portable and rapid diagnostic technology platform
peakPCR.